Chemical Structures in Databases (blog article 6 of 8) : Chemical Awareness
One of the many roles of IUPAC has been to establish and maintain systematic rules for the nomenclature of chemical entities. Within the chemical community these rules are simply referred to as IUPAC Nomenclature Rules and the systematic name for a chemical entity the IUPAC Name. Two excerpts taken verbatim from the 1993 recommendations (ISBN 0-632-03488-2) are i) “identify a chemical species by means of written or spoken words” and ii) “… important to recognise that the rules of systematic nomenclature need not necessarily lead to a unique name for each compound, but always lead to an unambiguous one”.
Perhaps two less well known IUPAC projects are IChIP (IUPAC Chemical Identifier Project) and InChI (IUPAC International Chemical Identifier). In contrast to the IUPAC name, these projects have examined the derivation of a unique digital name from a defined chemical structure. A goal is to use this identifier as a common language, perhaps even a standard, for the exchange of information about any given discrete compound in chemical database systems and/or supporting applications. The identifier is reversible, that is from a chemical structure the unique identifier can be generated and vice versa.
In May 2005, IUPAC published the source code and documentation for the generation of InChI identifiers from a discrete chemical structure. Both the InChI binaries and source code are available , and more recently also include molfile V3000 support.
Searching on InChI identifiers and SMILES strings
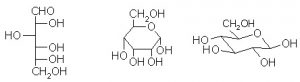
One of the most common user interfaces to a chemically enhanced DBMS represents allows chemical entities to be queried using SMILES strings. It is expected that COTS chemical structure database products will be enhanced to also support InChI identifiers. It is important to appreciate that, for example, a SMILES string represents the way a chemical entity is drawn. It should be further appreciated that even simple chemical entities can be drawn in a number of different ways. Given there is no unique rendition of a given entity, under the bonnet within the chemical database indexing component, the structure must be massaged, normalised and canonicalised in order to retrieve the expected database result set. For example, the structures below are all D-glucose. A query executed against a chemical structure database might be expected to retrieve the same database result set with different SMILES strings or InChI identifiers for glucose as drawn below (open chain Fischer projection, α- and β-anomers drawn in Haworth and 4C1 representations).
To reiterate, the chemical database extensible indexing component (eg. the Oracle Data Cartridge, IBM DB2 Extender) must understand the chemical significance of the way any chemical moiety is drawn, or more appropriately that the SMILES string, InChI identifier, or molfile used as a query predicate can be represented in more than one chemically significant and correct way.
Stereoisomerism
The InChI identifier for cis butene is “1/C4H8/c1-3-4-2/h3-4H,1-2H3/b4-3-“. If this identifier were stored in a database table, the SQL used to retrieve the database result set would be trivial. Given however that chemical database exact searches are only one type of search supported by most chemical database systems, the query is very unlikely to always be trivial. For example to query a chemical structure database where the cis but-2-ene component was a substructure of some larger chemical entity, a simple relational operation would not meet the requirements.Using database extensible indexing technology such as Oracle data cartridges, the application developer or DBA will most likely be exposed to a number of database operators for manipulation or query of chemical moieties. As few major chemical structure database vendors currently support InChI identifiers, the example below is a hypothetical SQL query using the database operator molExact (and not give free advertisement to the small handful of COTS chemical DB implementations). MolExact is an operator that returns a number greater than 0 (zero) should the exact chemical structure be found in the database, otherwise 0. This is consistent with other operators in chemical database systems outlined in COTS chemical structure database systems previously, and the contains operator briefly demonstrated on this website during the discussion of Oracle Text.
SELECT something
FROM tableName
WHERE molExact(columnName,'1/C4H8/c1-3-4-2/h3-4H,1-2H3/b4-3-')>0
The obvious needs to be clearly stated. Cis and trans butene are two very different compounds. Cis butene has a boiling point of 3.7°C and a melting point of -138.9°C; trans butene has a boiling point of 0.9°C and a melting point of -105.5°C. The connectivity tables however, represented internally as a type of adjacency matrix, are the same. Within the extensible indexing component of the chemical database, the system must be chemically aware and understand the differences between the two of them both despite identical adjacency matrices!
The SQL query above would only return information from the database relevent to cis butene. It would not retrieve information on trans butene or other isomers. Discussion on how this is achieved can be found elsewhere on this website. The same chemical awareness is required for other stereochemical properties and spatial bond arrangement around, for example, the tetrahedral carbon. This includes absolute configuration and the configuration of stereocentres relative to other parts of the same chemical entity.
Keto enol tautomerisation
In addition to stereoisomerism discussed briefly above, the extensible mechanism should understand other basic chemical principles such as keto enol tautomerism. The chemical structures below are all phenol. Although in practise a synthetic organic chemist would require good reason to query a chemical enhanced DBMS for phenol in a form other than its aromatic representation, regardless a database query using any of the tautomers as a search criteria should return identical database result sets.
This phenomenen of tautomerisation is also evident in 2-en-1-ols (the more common tautomer referred to as an aldehyde), enamines and imines etc.
Salt Stripping
Simply, salt stripping is a process of removing salts from a chemical entity. Searching a chemical structure database for the sodium salt of phenol, sodium phenolate, should allow results for phenol to be included in the database result set. In this case, phenol would also be included in the database result set for other salts such as potassium phenolate, lithium phenolate, or salts of other than simple alkali metals. The function of salt stripping permits the chemically enhanced DBMS to manipulate and/or retrieve information on the core chemical entity.
Salt stripping is a complex procedure. It is not always trivial to determine which of the components in a chemical moiety is a salt. In simple examples where alkali metals or mineral acids from amine salts are stripped, the methodology is clear; where the salt is for example a tartrate or protonated strychnine for example, things become clouded. Accordingly what constitutes a salt must often be predefined. The situation is further clouded with zwitterionic species such as amino acids and their salts. The chemically enhanced database should permit the same database result set to be generated irrespective of the search term representing the chemical moiety. The extensible indexing mechanism and supporting routines should provide functionality for salt stripping.
Radio labelling, isotopic labelling, or isotopic depletion
No tetravalent carbon atom can ever be dismissed as just a carbon atom, for example. It may be 13C enriched, or even 13C depleted or labelled in other ways (14C) for a number of reasons, not least that include the determine of some chemical mechanistic pathway. It is not an unreasonable request for a chemical database query to be expected to satisfy not only atom-atom connectivity expressed as an adjacency matrix, or bond order, or valency, but isotopic labelling too. Although this task can be performed in database result set post processing step, this removes the ability to chemically query a database using the language of the (relational) database, SQL. Any respectful chemical RDMBS implementation should be chemically aware at RDBMS level, and not using some bespoke external API that was implemented in a custom library or middleware software.
Miscellaneous
Even simple functional groups such as azides, nitro groups, S– and N-oxides must be handled with care. The extensible indexing component must normalise these functional groups, and the whole molecule or substructure, during the creation and updating of the underlying database indexes and also prior to querying/searching these indexes. This task is often both computationally expensive and may require a second “pass” of the intermediary result-set to afford the desired database result-set.
Summary
Querying a chemical structure database is not as trivial as generating some unique identifier from a given chemical structure followed by a few relational operations. A set of database operators or functionality encapsulated within database stored procedures or external routines will be defined for any extensible indexing component. This functionality parlays much chemical awareness to handle stereoisomerism, keto enol tautomerisation, mutarotation, salt stripping, and zwitterionic species appropriately. The molExact operator defined above, or the three operators demonstrated on this website previously and supporting library routines, encapsulate much of this functionality.
This is the sixth of eight blog articles with a subtitle of “Chemical Structures In Databases”. The prior article can be located here and the next article here.
— Published by Mike, 21:27:09 26 September 2018 (BST)
Archive
By Month: November 2022, October 2022, August 2022, February 2021, January 2021, December 2020, November 2020, March 2019, September 2018, June 2018, May 2018, April 2018
Categories
Apple, C#, Databases, Faircom, General IT Rant, German, Informatics, LINQ, MongoDB, Oracle, Perl, PostgreSQL, SQL, SQL Server, Unit Testing, XML/XSLT



Leave a Reply