Chemical Structures in Databases (blog article 5 of 8) : Subgraph Isomorphism
A mathematician might define subgraph isomorphism as the determination of a subgraph of a graph which is isomorphic to another graph. From the chemo-informatic practitioners perspective, specifically relating to 2D chemical structure (the atom-atom connectivity in a chemical structure is a graph after-all, albeit in disguise) databases, the definition might be more loosely defined as a chemical substructure search.
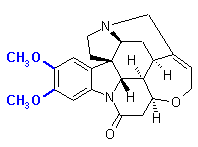
A typical substructure search might involve retrieving all compounds from a from a chemical database that look like strychnine. The action of executing a query of this type results in many compounds being included in the database result set, including brucine (the differences between strychnine and brucine highlighted in blue).
The hypothetical database query shown (pseudo SQL) is an example of graph/subgraph isomorphism applied to chemoinformatics. To retrieve the alkaloid brucine using this type of SQL, a while wildcard match for strychnine would need to be introduced, using a LIKE % clause and R-groups on the aromatic ring.
Out of the box relational databases such DB2, Oracle, SQL Server, PostgreSQL and so on are not chemically aware. The ANSI/ISO recommendations do not describe, and the vendor database implementations do not offer, enhanced SQL with functionality to perform, for example, chemical structure (exact) or substructure searches. For DBMS implementations that require chemical awareness, the void is addressed with third party extensible indexing database components. The COTS products of various informatics provides offer solutions to address this void.
Within the extensible indexing component, chemical entities can be represented as adjacency matrices. For example, the vertex-adjacency matrix A(G) of a labelled connected graph G (urea) with 4 vertices (excluding hydrogen) is the 4 (rows) x 4 (columns) diagonal matrix shown.
To perform a substructure search, that is to determine whether strychnine contains a substructure with the atom connectivity of urea, a database plug-in responsible for the indexing and manipulation of chemical entities would have to find an occurrence of the adjacency matrix A(G) in the similarly constructed adjacency matrix for strychnine. The graph atom connectivity observed in urea is not in strychnine; searching a chemical structure database for all compounds containing the connectivity of urea (excluding hydrogen atoms) should not retrieve strychnine in the database result set. If a chemical structure database were searched for information about chemical entities that contained the atom connectivity exhibited by indole as a substructure however, strychnine would be included in the database result set. In this instance, the chemoinformatics practitioner is querying the chemical structure database for all moieties that contain the substructure of indole.
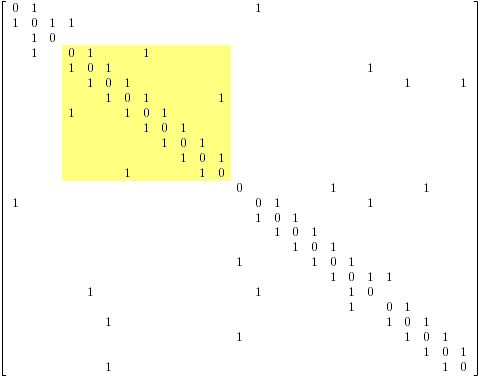
The two adjacency matrices A(Indole1) and A(Indole2) above both represent adjacency matrices for indole. There are others; many others. The chemically aware indexing component will search the adjacency matrix of strychnine for the occurrence of the adjacency matrix of indole. The connectivity observed for indole and represented in its adjacency matrix A(Indole1) is present in the adjacency matrix for strychnine A(Strychnine).
When substructure searches are performed, or when key index fields required for SQL predicates as described in the Cope-Chat discussion are required, comparisons of adjacency matrices are likely to be performed. For example, to confirm that strychnine possesses the substructure index keys (eg. 3o amine, allylic ether, amide, indole, amide, number and size of rings, chiral flag), or to confirm that the substructure of strychnine is contained within brucine.
It should be noted that two different indole adjacent matrices for indole are shown above. The second adjacency matrix differs from the first in that the atom numbering has been slightly varied. In this example the detection of the alternative (the one without the yellow background) within A(Strychnine) is not trivial. The atom numbering in A(Strychnine), A(Indole1), and A(Indole2) is arbitrary. Variation in the numbering of any of the graphs may result in adjacency matrices other than those shown herein. Accordingly the detection of A(Indole2) in A(Strychnine) may require considerable computational resource. Such tasks however need not require expensive resource if sensible algorithms are used (ie. specifically not some silly combinatorial brute force approach – as recently as two days ago Jarek Duda published preliminary details of graph isomorphism detection in polynomial time).
Adjacency Matrix Variations
The adjacency matrix is one of a number of graph theoretical matrices that can be used for
manipulation of chemical structure data. Other variations useful for chemical structure databases are the distance matrix, or vertex and edge-weighted matrices encompassing the vertex label (the chemical element) and bond type (sp3, sp2, and sp, tautomeric, aromatic, ionic etc).
References
The reader is directed to the seminal work of Ullman (Ullman, J. R.; An Algorithm for Subgraph Isomorphism, J. ACM, vol. 23(1) 1976) for further discussion and citation references. At the time of writing (September 2018), Professor Brendan D. McKay claims that nauty is the world’s fastest isomorphism testing implementation. Further information including C source code can be found on the nauty website. Readers are also directed to the VFLib Graph Matching Library, a C++ implementation of a number of graph matching algorithms. An implementation of this algorithm is also available in C# from CodeProject.
Summary
If database indexes were to be implemented for chemical moiety functional groups, as outlined during the discussion of Cope-Chat cards, or comparison of the structure of one graph represented as an adjacency matrix with another within the extensible indexing component, graph and subgraph isomorphism detection would be used. The implementation of these algorithms would be the crux of the chemical structure database extensible indexing component. In Oracle, these components are referred to as Oracle Data Cartridges [or better put a technical implementation enabling Oracle to index a custom data type exist not only for chemical structures, but human (and cats, horses, and extraterrestrial aliens) DNA profiles as implemented in C$WILDNA1 for example].
The underlying theory describing subgraph isomorphism is described in the primary literature. The source code for some some subgraph isomorphism implementations are available in the public domain, and open source.
COTS products that encapsulate structure and substructure searching within chemical structure database systems are available.
This is the fifth of eight blog articles with a subtitle of “Chemical Structures In Databases”. The prior article can be located here and the next article here.
— Published by Mike, 16:38:43 22 September 2018 (BST)
Archive
By Month: November 2022, October 2022, August 2022, February 2021, January 2021, December 2020, November 2020, March 2019, September 2018, June 2018, May 2018, April 2018
Categories
Apple, C#, Databases, Faircom, General IT Rant, German, Informatics, LINQ, MongoDB, Oracle, Perl, PostgreSQL, SQL, SQL Server, Unit Testing, XML/XSLT







Leave a Reply