MongoDB as a DNA Typing Database (SGMPlus?)
Is MongoDB up to the job of being able to query a database of ca. 10,000,000 SGM Plus DNA profiles and laboratory samples? Do I have to carefully consider JSON document design? Is the database performant? I need to know the answer to these questions, and more.
I have undertaken many projects with a DNA database focus. Last month I documented some introductory findings in a small blog post on MongoDB. A month later, I retain my interest in MongoDB. I need however to prove to myself that MongoDB isn’t a toy or fad and that it can cut the mustard when used in a typical informatics system. A reasonably sized SGMPlus DNA database is a good test case to explore application of MongoDB, and it is a domain where I have prior experience and knowledge. Having performed these MongoDB evaluations offline, I have decided up write up this blog article with some of my findings, and include some code snippets too for any readers that might choose to use the content herein for something useful. Before jumping straight in and copy/pasting the source code however, I need to put things into context given not everyone reading my blog will be familiar with the subject matter. The structure of this blog article will be:
- Some background introduction to the basics of DNA typing, the key experimental steps undertaken within the laboratory, how samples are tracked through the laboratory workflow, and importantly what an SGMPlus profile might look like. The coverage is far from thorough and limited to the detail necessary to demonstrate the applicability of MongoDB for usage in a DNA typing database. I have also attempted to use layman terms where I can to keep the focus on MongoDB.
- Generation of test data representing some laboratory sample tracking information, and SGMPlus DNA profiles. The test data is produced with a little Perl and represented, for ingestion into MongoDB, as an array of JSON documents.
- Show that MongoDB queries can be formulated to address the typical types of queries executed against a DNA database.
Background: The Sample And Its Workflow Through The Laboratory
After a sample has been delivered to the forensics laboratory, it is examined prior to, possibly, undergoing some wet chemistry. Some of the typical steps in this workflow are summarized below, and this information maps directly to the fields in a JSON representation of the workflow process, and also not atypical database queries performed against an SGMPlus DNA database. The same general processes will occur for other multiplex kits too, eg. SGM, Y-Filer, NGM, and Minifiler.
These workflow processes are summarized below, but in-advance the general workflow is: Sample (eg. from a crime scene) Examination → Sample DNA Extraction (→ optional DNA Quantitation) → PCR Fragment Amplification → Gel or Capillary Electrophoresis → Data Analysis and DNA Profile Designation → send it off and raise the invoice.
Sample Examination
Upon arrival within the (forensic) laboratory, the sample (or samples all from the same case or project) is unpackaged, examined, and a decision made on:
- Whether the sample is a good candidate for DNA typing: Is DNA more likely to be obtained from a knife covered in blood, semen from a vaginal swab, or a Buccal swab over, for example, a potential sweat stain on a cola bottle found at the crime scene.
- Commercial implications and budgets: is it worthwhile to examine, swab, and attempt DNA detection from scores of samples over one or two samples for only a minor offence.
- SLA’s: Given a suspect can be held for only a limited time in a police cell or as instructed by the courts, evidential samples that have the fastest throughput or that are routine may be favoured over other samples.
There are of course many other considerations that are taken into account during sample examination, eg. sample contamination or age, but the point is that the sample is examined and it may or may not be selected as a candidate to be evaluated further within the laboratory.
If the sample is selected, it may undergo a wet chemistry step to extract the DNA via. an extraction process.
Sample DNA Extraction
DNA extraction is the wet chemistry process whereby the DNA is extracted from the other material. The process itself will depend on the type of sample being examined. For example, extracting DNA from (white) blood cells, sperm, hair roots, hair shafts, sweat/touch/saliva residues (eg. envelope sealing flaps, cigarette butts, chewing gum, and drink bottles), and bones will all require a different experimental process for DNA extraction. One of the most common methods for obtaining a DNA sample under controlled conditions from a suspect by UK police is through a Buccal swab; a few cells collected from the inside cheek by scraping with a mildly abrasive surface. The photograph below shows former UK Prime Minister Tony Blair voluntarily receiving a Buccal Swab by Detective Constable Jayne Thomas at Medway Police Station in Kent UK. He was neither charged nor convicted of any offence.
These processes for DNA extraction are well documented in organization SOP and IOP’s, and very often also under regulatory control. DNA extracted other sample types, including aged or degraded bones, decomposed body carcasses etc. may require specialist expertise using atypical methodologies. Overall however, the basic process is that within the laboratory, the sample is extracted through one of a number of processes, cells containing DNA (nuclear or perhaps even mitochondrial) are broken down, their DNA released, and an aliquot of the liquid fraction containing the DNA separated from the remaining sample material. For automated processes, the sample containing the potentially extracted DNA (see quantitation) will deposited via pipette onto a plate, and its location recorded. Diagrammatically in the 96 well plate below, plate number E123456, a sample is being deposited via. pipette in well location H06.
Other plate sizes exist too, notably the 384 (4 * 96) well plate, with defined well locations A01 to F24, and the 1536 well plate (16 * 96 or 4 * 384, depending on your perspective).
DNA Quantitation
Quantitation is the experimental process use to determine the existence of DNA, and if present, to estimate the amount present. The general technologies used range from a simple colourimetric reactions and staining to automated (ie. laboratory robotics) quantitation using LC fluorescence spectrometers. Sometimes too the techniques detect human genomic DNA specifically, over the total DNA load that will include DNA from other sources (eg. bacteria or animals). Although the quantitation process is generally not lossful, more often than not the small sample aliquot is not recovered from the process to use in any subsequent PCR amplification step.
After quantitation, the extraction samples often undergo a process known as plate reformatting, that in layman terms means moving the samples from the extraction plate, to another plate, for PCR amplification. Extraction samples that are proven to not contain DNA, ie. quantitation shows no or little DNA present, are not transferred on to the PCR plate; extraction samples containing DNA are transferred through to the PCR plate for subsequent reactions, often diluted or concentrated, to the optimal amount required for PCR.
Quantitation may or may not involve a different plate to the extraction plate, however the point I need to make is that it is likely. Put another way, should the samples in extraction plate number E12346 well location E05, and plate number E12345 well locations H07 and H08 be shown to respectively contain, contain, and not contain DNA, two samples will be remapped to another plate (eg. P654321) and one sample (plate E12345 well location H08) discarded (it contains no DNA, what is the point of processing it further and at cost?).
PCR (Polymerase Chain Reaction) DNA Fragment Amplification
Modern COTS fast high throughput DNA profiling may use PCR as the key DNA amplification step. The background to this technology, that won chemist (of course) Kary Mullis the Chemistry Nobel Prize in 1994, is readily available over the internet, the primary literature, and in readable textbooks such as John Butler’s Forensic DNA Typing (ISBN-13: 978-0121479527) and I will not attempt to dumb down the technology and technique in a paragraph or two. In summary however, PCR is a proven technology that will enable rapid amplification of a very specific region of DNA multifold, where multi is generally a large number. For the purposes of human identification and DNA typing, these very specific regions have a different repeating pattern, an STR in the jargon, that will enable the DNA of one person to be differentiated from another person, more-so if multiple of these regions are amplified (and detected).
Gel (or Capillary Electrophoresis)
Capillary electrophoresis is specialised form of chromatography often used as a separation method in DNA analysis. With forensic application, the process can be viewed as a routine fast turn-around automated technology used to separate, detect, and quantify individual (fluorescently) labeled DNA fragments (from PCR amplification in the prior wet-chemistry step).
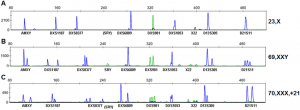
One typical piece of CE automation technology is an ABI PRISM 3700 Genetic Analyzer, and its powerful processing software for DNA profile analysis ultimately used to designate DNA profiles. A resultant typical chromatogram trace (containing some loci designations) is shown below.
DNA Profile Designation (Data Interpretation)
DNA profile designation is the output of DNA profiling, and the step following the Gel/CE. This step in the workflow can be summarized as competent individuals confirming that from all the laboratory processes, the DNA profile produced has come from the sample provided to the laboratory (and represents a single individual and full, partial, or mixed profile). The profile itself are loci (labels used to identify a published position/location on a chromosome, eg. vWA, TH01, FGA) and some numbers that represent the number of times a short pattern of bases are repeated (STR). The loci are chosen to be highly discriminatory, ie. the number of the repeating blocks will likely be different between us (you the reader and someone else).
Here is my SGMPlus DNA profile for example.
vWA (13,2, 18.2)
D18 (10, 21.2)
D2 (19.2, 25.2)
TH01 (5.1, 9.3)
FGA (28, 28)
D3 (19, 21)
D21 (26, 32.1)
D16 (9.2, 14)
D19 (8, 8)
D8 (14, 17)
Amel (X, Y)
Other multiplexes/profiling kits will result in DNA profiles that look subtly different. The loci set for Y-Filer & CODIS for example are not the same as for SGMPlus.
This is the SGMPlus profile that would result from me leaving my cellular material around a crime scene, so you or both of us would have a good chance of getting away with premeditated murder for example. Note clearly this isn’t my human genome, just sufficient information produced quickly using automated techniques that will enable me (my DNA) to be differentiated from yours, and most likely everyone else in your country, and probably everyone else in the world. Think of it like this – I am a man, if I committed a crime and my DNA was found at the crime scene, around half the worlds population can excluded immediately (they are women, Amel/Amelogen is the marker used to indicate genetic sex). For another loci, where the value pair is not binary (viz. XX or XY) but two of 20 or 30, the technique becomes exceptionally discriminatory very quickly. Note too that there are 10 loci/markers (and sex/Amel), so it is easy to comprehend that this is a very discriminatory technique for matching DNA samples.
Informatics: The laboratory wet chemistry workflow Sample Tracking and SGM Plus DNA Profile
Having painted a backdrop to DNA typing, plate and well locations and plate reformatting, I can now define two not unrealistic JSON documents that could represent the sample plate ID’s and well locations, a numeric forensic case identifier, and perhaps SGM Plus profile, for some arbitrary sample.
The first JSON document shows the sample ID, some unique identification number to differentiate and track this specific sample through the laboratory and reporting processes, and details of the extraction and quantitation plates (and well locations on the plate, adhering to the plate coordinates discussed above). Notably no PCR or other wet chemistry processes occurred (in my contrived example data, extraction plates are prefixed with an E, quantitation plates a Q, PCR places a P, and capillary/electrophoresis places a C); perhaps, as discussed previously, the quantitation step confirmed that the sample under investigation contained no or insufficient quantities of DNA?
{
"sampleID" : 1,
"plateNumberEXT" : "E00032461",
"wellLocationEXT" : "D05",
"plateNumberQUANT" : "Q00005500",
"wellLocationQUANT" : "B09",
"caseID" : 472977
}
The second JSON document shows another sample with complete sample tracking through the extraction, quantitation, PCR, and CE/Gel process, and a designated SGMPlus profile that also exhibits trisomy on chromosome 16, a homozygous 23, 23 designation for FGA (chromosome 4), allelic designations for 10 autosomal markers and a designation for of X, F for amelogenin (amelogenin is a gene that codes for a protein observed in tooth enamel on both the X and Y chromosomes, and subtle sequence length differences in the amelogenin gene on the X and Y chromosomes are a good first guess to differentiate males from females; one allele is designated X and the other as F=unknown/fail/cannot determine etc). Importantly too, the allelic designations are not just random numbers and letters, they are a quantised set of allowed values that correspond to the most common alleles for each locus/marker for the DNA profile multiplex (ie. SGMPlus). For the purposes of QA/QC, reporting, and occasionally informatics purposes, the allowed allelic count for each locus must also marry up with an allelic ladder.
{
"sampleID" : 2,
"plateNumberEXT" : "E00032461",
"wellLocationEXT" : "D06",
"plateNumberQUANT" : "Q00005500",
"wellLocationQUANT" : "B10",
"plateNumberPCR" : "P00021200",
"wellLocationPCR" : "D02",
"plateNumberCE" : "C00017800",
"wellLocationCE" : "E12",
"profile" : [
{ "locus" : "TH01" , "alleles" : ["6.3","8.1"] },
{ "locus" : "FGA" , "alleles" : ["23, 23"] },
{ "locus" : "D21" , "alleles" : ["24.3","35.1"] },
{ "locus" : "Amel" , "alleles" : ["F","X"] },
{ "locus" : "D18" , "alleles" : ["25.2","12"] },
{ "locus" : "D8" , "alleles" : ["8.2","6"] },
{ "locus" : "D3" , "alleles" : ["15.2","18"] },
{ "locus" : "D19" , "alleles" : ["11.2","7"] },
{ "locus" : "D2" , "alleles" : ["25","21"] },
{ "locus" : "vWA" , "alleles" : ["23","10"] },
{ "locus" : "D16" , "alleles" : ["11","12.2","12"] }
],
"caseID" : 171162
}
Obtaining real population DNA profile data is notoriously difficult, not least because jurisdictional laws prohibit disclosure and/or distribution except under all but very controlled situations [the data is very frequently obtained as part of criminal justice investigations (eg. involving for example murder and rape), civil action (eg. proof of sirehood and alimony obligations), human identity (evidential proof of the identity of Saddam and Uday Hussein, or Osama bin Laden)], and sometimes even commercial implications.
For the purposes of evaluating whether MongoDB had the necessary feature set and capacity to handle a large database containing laboratory workflow details and SGMPlus DNA profiles, actual data for 10,000,000 samples is not necessary. A script to generate example data would more than suffice, as long as it approximated the allelic designations etc.
The Perl script below generates this example data (this is a modification of a Perl script I have previously written and published for the purposes of C$WILDNA1 performance testing. I have also previously formatted SGM Plus DNA profile excerpt data with C# and LINQ).
#!/usr/bin/perl -w
use POSIX qw(ceil);
#initial dummy variable values for plate details
my @labPlateSerialNo = (32461,5500,21200,17800);
my @labPlateSerialSuffix = ("EXT","QUANT","PCR","CE");
my @labOperationProb = (1,0.95,0.9,0.9);
my @labPlateWellLoc = (41,21,50,76);
my @labPlateRows = (8,8,24,24);
my @labPlateCols = (12,12,16,16);
my @sgmPlus=( # marker low high
# ------- ------------------------------------------------------- ---------------------------------------------------------------
[ "D2", [14..29,15.2,16.2,17.2,18.2,19.2,20.2,21.2,22.2,23.2,
24.2,25.2,26.2,27.2,F], [14..29,15.2,16.2,17.2,18.2,19.2,20.2,21.2,22.2,23.2,24.2,
25.2,26.2,27.2]],
[ "D3", [9..21,12.2,13.2,14.2,15.2,16.2,17.2,18.2,F], [9..21,12.2,13.2,14.2,15.2,16.2,17.2,18.2,F]],
[ "D8", [6..20,8.2,9.2,10.2,11.2,12.2,13.2,14.2,15.2,
16.2,17.2,18.2,F], [6..20,8.2,9.2,10.2,11.2,12.2,13.2,14.2,15.2,16.2,17.2,
18.2,F]],
[ "D16", [4..16,5.2,6.2,7.2,8.2,9.2,10.2,11.2,12.2,13.2,14.2,F], [4..16,5.2,6.2,7.2,8.2,9.2,10.2,11.2,12.2,13.2,14.2,F]],
[ "D18", [6..28,9.2,10.2,11.2,12.2,13.2,14.2,15.2,16.2,17.2,
18.2,19.2,20.2,21.2,22.2,23.2,24.2,25.2,26.2,F], [6..28,9.2,10.2,11.2,12.2,13.2,14.2,15.2,16.2,17.2,18.2,
19.2,20.2,21.2,22.2,23.2,24.2,25.2,26.2,F]],
[ "D19", [7..17,9.2,10.2,11.2,12.1,12.2,13.2,14.2,15.2,16.2,
17.2,18.2,F], [7..17,9.2,10.2,11.2,12.1,12.2,13.2,14.2,15.2,16.2,
17.2,18.2,F]],
[ "D21", [23..39,24.2,24.3,25.2,26.2,27.2,28.1,28.2,28.3,29.1,
29.2,29.3,30.1,30.2,30.3,31.1,31.2,31.3,32.1,32.2,
32.3,33.1,33.2,33.3,34.1,34.2,34.3,35.1,35.2,35.3,
36.2,37.2,F], [23..39,24.2,24.3,25.2,26.2,27.2,28.1,28.2,28.3,29.1,29.2,
29.3,30.1,30.2,30.3,31.1,31.2,31.3,32.1,32.2,32.3,33.1,
33.2,33.3,34.1,34.2,34.3,35.1,35.2,35.3,36.2,37.2,F]],
[ "Amel", [X,F], [X,Y,F]],
[ "FGA", [16..33,42..51,17.2,18.2,19.2,20.2,21.2,22.2,23.2,
24.2,25.2,26.2,27.2,28.2,29.2,30.2,31.2,32.2,33.2,
42.2,43.2,44.2,45.2,46.2,47.2,48.2,50.2,51.2,F], [16..33,42..51,17.2,18.2,19.2,20.2,21.2,22.2,23.2,24.2,
25.2,26.2,27.2,28.2,29.2,30.2,31.2,32.2,33.2,42.2,
43.2,44.2,45.2,46.2,47.2,48.2,50.2,51.2,F]],
[ "TH01",[3..13,5.1,5.3,6.1,6.3,7.1,7.3,8.1,8.3,9.1,9.3,13.3,F], [3..13,5.1,5.3,6.1,6.3,7.1,7.3,8.1,8.3,9.1,9.3,13.3,F]],
[ "vWA", [9..25,11.2,12.2,13.2,14.2,15.2,16.2,17.2,18.2,
19.2,20.2,21.2,22.2,23.2,24.2,F], [9..25,11.2,12.2,13.2,14.2,15.2,16.2,17.2,18.2,
19.2,20.2,21.2,22.2,23.2,24.2,F]]
);
#shuffle a 1D array of numbers
sub shuffle1D
{
my $aOfN=shift;
for (my $i= @$aOfN;--$i;)
{
my $j = int rand($i+1);
@$aOfN[$i,$j] = @$aOfN[$j,$i];
}
}
#returns random array element from 1D array
sub rnElement
{
my $aOfN=shift;
return @$aOfN[int rand(scalar(@$aOfN))];
}
sub createDNAprofile
{
my @locusOrder=(0..$#sgmPlus);
shuffle1D(\@locusOrder);
print " \"profile\" : [\r\n";
for (my $j=0;$j<scalar(@locusOrder);$j++) { my $sgmIndex = $locusOrder[$j]; #minimum of a value for one chromosome loci (monosomy) #disomy normal/common, other aneuploidy possible althrough rare and accommodating #for trisomy, tetrasomy, and pentasomy my $one = rnElement($sgmPlus[$sgmIndex][1]); my $two = rand(100)>5?rnElement($sgmPlus[$sgmIndex][2]):"";
my $three = rand(100)>98?rnElement($sgmPlus[$sgmIndex][2]):"";
my $four = rand(100)>98?rnElement($sgmPlus[$sgmIndex][2]):"";
my $five = rand(100)>98?rnElement($sgmPlus[$sgmIndex][2]):"";
my $desigList = join ',', map qq("$_"), grep { length } ($one, $two, $three, $four, $five);
print " { \"locus\" : \"$sgmPlus[$sgmIndex][0]\" , \"alleles\" : [$desigList] }".terminalChar(1+$j,scalar(@locusOrder))."\r\n";
}
print " ],\r\n";
}
sub wellLocation
{
my $rows=shift;
my $cols=shift;
my $locationNumeric=shift;
my $col = ($locationNumeric-1)%$cols +1;
my $row = ceil(($locationNumeric % ($rows*$cols+1)) / $cols);
return sprintf("%c%02d", $row+64,$col);
}
#recursively generate extraction, quantitation, pcr, and gel/ce plate information for the lab operation
sub labOperation
{
my $opType=shift;
my $probabilityOfFailure = $labOperationProb[$opType];
if (rand()<=$labOperationProb[$opType]) { printf(" \"plateNumber%s\" : \"%s%08d\",\r\n",$labPlateSerialSuffix[$opType],substr($labPlateSerialSuffix[$opType],0,1),$labPlateSerialNo[$opType]); printf(" \"wellLocation%s\" : \"%s\",\r\n",$labPlateSerialSuffix[$opType],wellLocation($labPlateRows[$opType],$labPlateCols[$opType],$labPlateWellLoc[$opType])); #increment/reset well location, and/or increment the plate serial number if ($labPlateWellLoc[$opType]>=($labPlateRows[$opType]*$labPlateCols[$opType]))
{
$labPlateWellLoc[$opType] = 0;
$labPlateSerialNo[$opType]++;
}
$labPlateWellLoc[$opType]++;
if ($opType<3)
{
labOperation(++$opType);
}
else { createDNAprofile }; #so if a gel/capillary electrophoresis lab operation performed, assume a DNA profile came out of it
}
}
#in a list, have commas after all items except the terminal char
sub terminalChar
{
my $currentIndex=shift;
my $terminalIndexPosn=shift;
return $currentIndex<$terminalIndexPosn?',':"";
}
my @locusOrder=(0..$#sgmPlus);
if ($#ARGV != 0 ) {
print "usage: generateSGM+.pl number, eg. \"generateSGM+.pl 3\" will generate 3 dummy DNA lab workflow JSON documents (as an array)\r\n";
exit;
}
my $noSamples = shift; #the number of SGM+ lab workflow JSON documents to generate
print "[\r\n";
for $sampleID (1..$noSamples)
{
shuffle1D(\@locusOrder);
print " {\r\n";
if (int(rand(1000)) == 500)
{
#Just spit out the DNA profile for about 1 in every 1000 without any of the laboratory workflow.
#This is not an atypical scenario for DNA profiles that have been imported from another laboratory/company, or for
#samples that are for example Police Elimination profiles. The lab profiles and other traceable information
#is not provided
createDNAprofile();
}
else
{
print " \"sampleID\" : $sampleID,\r\n";
labOperation(0);
}
printf(" \"caseID\" : %d\r\n", int(rand(1000000)));
printf(" }%s\r\n",terminalChar($sampleID,$noSamples));
}
print "]\r\n";
The script above was used to generate the 10,000,000 samples, the vast majority that also contained SGM Plus profile JSON data. The complete log for profile generation, and import into MongoDB, is shown below.
Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ ./generateSGM+.pl 10000000 > sgmplusTestData.JSON Michaels-MBP:json sgm+ mdo$ Michaels-MBP:json sgm+ mdo$ ls -s sgmplusTestData.JSON 17662080 sgmplusTestData.JSON Michaels-MBP:json sgm+ mdo$ mongoimport --db=mikedb --collection=sgmplus --jsonArray --file=sgmplusTestData.JSON 2017-11-26T16:34:16.100+0100 connected to: localhost 2017-11-26T16:34:19.090+0100 [........................] mikedb.sgmplus 84.4MB/8.41GB (1.0%) 2017-11-26T16:34:22.093+0100 [........................] mikedb.sgmplus 170MB/8.41GB (2.0%) 2017-11-26T16:34:25.092+0100 [........................] mikedb.sgmplus 256MB/8.41GB (3.0%) 2017-11-26T16:34:28.095+0100 [........................] mikedb.sgmplus 342MB/8.41GB (4.0%) 2017-11-26T16:34:31.095+0100 [#.......................] mikedb.sgmplus 427MB/8.41GB (5.0%) 2017-11-26T16:34:34.090+0100 [#.......................] mikedb.sgmplus 513MB/8.41GB (6.0%) 2017-11-26T16:34:37.094+0100 [#.......................] mikedb.sgmplus 600MB/8.41GB (7.0%) 2017-11-26T16:34:40.090+0100 [#.......................] mikedb.sgmplus 684MB/8.41GB (7.9%) 2017-11-26T16:34:43.091+0100 [##......................] mikedb.sgmplus 769MB/8.41GB (8.9%) 2017-11-26T16:34:46.095+0100 [##......................] mikedb.sgmplus 851MB/8.41GB (9.9%) 2017-11-26T16:34:49.092+0100 [##......................] mikedb.sgmplus 935MB/8.41GB (10.9%) 2017-11-26T16:34:52.095+0100 [##......................] mikedb.sgmplus 1016MB/8.41GB (11.8%) 2017-11-26T16:34:55.090+0100 [###.....................] mikedb.sgmplus 1.08GB/8.41GB (12.8%) 2017-11-26T16:34:58.091+0100 [###.....................] mikedb.sgmplus 1.16GB/8.41GB (13.8%) 2017-11-26T16:35:01.095+0100 [###.....................] mikedb.sgmplus 1.25GB/8.41GB (14.8%) 2017-11-26T16:35:04.094+0100 [###.....................] mikedb.sgmplus 1.33GB/8.41GB (15.8%) 2017-11-26T16:35:07.090+0100 [####....................] mikedb.sgmplus 1.41GB/8.41GB (16.8%) 2017-11-26T16:35:10.093+0100 [####....................] mikedb.sgmplus 1.49GB/8.41GB (17.7%) 2017-11-26T16:35:13.094+0100 [####....................] mikedb.sgmplus 1.57GB/8.41GB (18.6%) 2017-11-26T16:35:16.095+0100 [####....................] mikedb.sgmplus 1.64GB/8.41GB (19.6%) 2017-11-26T16:35:19.095+0100 [####....................] mikedb.sgmplus 1.72GB/8.41GB (20.5%) 2017-11-26T16:35:22.094+0100 [#####...................] mikedb.sgmplus 1.80GB/8.41GB (21.4%) 2017-11-26T16:35:25.095+0100 [#####...................] mikedb.sgmplus 1.88GB/8.41GB (22.3%) 2017-11-26T16:35:28.092+0100 [#####...................] mikedb.sgmplus 1.95GB/8.41GB (23.2%) 2017-11-26T16:35:31.095+0100 [#####...................] mikedb.sgmplus 2.03GB/8.41GB (24.2%) 2017-11-26T16:35:34.095+0100 [######..................] mikedb.sgmplus 2.11GB/8.41GB (25.1%) 2017-11-26T16:35:37.093+0100 [######..................] mikedb.sgmplus 2.18GB/8.41GB (26.0%) 2017-11-26T16:35:40.093+0100 [######..................] mikedb.sgmplus 2.26GB/8.41GB (26.9%) 2017-11-26T16:35:43.093+0100 [######..................] mikedb.sgmplus 2.34GB/8.41GB (27.8%) 2017-11-26T16:35:46.094+0100 [######..................] mikedb.sgmplus 2.41GB/8.41GB (28.7%) 2017-11-26T16:35:49.093+0100 [#######.................] mikedb.sgmplus 2.49GB/8.41GB (29.6%) 2017-11-26T16:35:52.091+0100 [#######.................] mikedb.sgmplus 2.56GB/8.41GB (30.5%) 2017-11-26T16:35:55.093+0100 [#######.................] mikedb.sgmplus 2.64GB/8.41GB (31.4%) 2017-11-26T16:35:58.093+0100 [#######.................] mikedb.sgmplus 2.72GB/8.41GB (32.3%) 2017-11-26T16:36:01.094+0100 [#######.................] mikedb.sgmplus 2.79GB/8.41GB (33.2%) 2017-11-26T16:36:04.094+0100 [########................] mikedb.sgmplus 2.87GB/8.41GB (34.1%) 2017-11-26T16:36:07.094+0100 [########................] mikedb.sgmplus 2.95GB/8.41GB (35.1%) 2017-11-26T16:36:10.093+0100 [########................] mikedb.sgmplus 3.02GB/8.41GB (36.0%) 2017-11-26T16:36:13.090+0100 [########................] mikedb.sgmplus 3.10GB/8.41GB (36.9%) 2017-11-26T16:36:16.094+0100 [#########...............] mikedb.sgmplus 3.18GB/8.41GB (37.8%) 2017-11-26T16:36:19.092+0100 [#########...............] mikedb.sgmplus 3.26GB/8.41GB (38.8%) 2017-11-26T16:36:22.094+0100 [#########...............] mikedb.sgmplus 3.34GB/8.41GB (39.7%) 2017-11-26T16:36:25.093+0100 [#########...............] mikedb.sgmplus 3.41GB/8.41GB (40.6%) 2017-11-26T16:36:28.093+0100 [#########...............] mikedb.sgmplus 3.49GB/8.41GB (41.5%) 2017-11-26T16:36:31.090+0100 [##########..............] mikedb.sgmplus 3.57GB/8.41GB (42.4%) 2017-11-26T16:36:34.090+0100 [##########..............] mikedb.sgmplus 3.64GB/8.41GB (43.3%) 2017-11-26T16:36:37.094+0100 [##########..............] mikedb.sgmplus 3.72GB/8.41GB (44.3%) 2017-11-26T16:36:40.090+0100 [##########..............] mikedb.sgmplus 3.80GB/8.41GB (45.2%) 2017-11-26T16:36:43.092+0100 [###########.............] mikedb.sgmplus 3.88GB/8.41GB (46.1%) 2017-11-26T16:36:46.090+0100 [###########.............] mikedb.sgmplus 3.95GB/8.41GB (47.0%) 2017-11-26T16:36:49.094+0100 [###########.............] mikedb.sgmplus 4.03GB/8.41GB (47.9%) 2017-11-26T16:36:52.094+0100 [###########.............] mikedb.sgmplus 4.11GB/8.41GB (48.8%) 2017-11-26T16:36:55.094+0100 [###########.............] mikedb.sgmplus 4.18GB/8.41GB (49.7%) 2017-11-26T16:36:58.094+0100 [############............] mikedb.sgmplus 4.26GB/8.41GB (50.7%) 2017-11-26T16:37:01.093+0100 [############............] mikedb.sgmplus 4.34GB/8.41GB (51.6%) 2017-11-26T16:37:04.094+0100 [############............] mikedb.sgmplus 4.42GB/8.41GB (52.5%) 2017-11-26T16:37:07.094+0100 [############............] mikedb.sgmplus 4.49GB/8.41GB (53.4%) 2017-11-26T16:37:10.094+0100 [#############...........] mikedb.sgmplus 4.57GB/8.41GB (54.3%) 2017-11-26T16:37:13.090+0100 [#############...........] mikedb.sgmplus 4.65GB/8.41GB (55.3%) 2017-11-26T16:37:16.091+0100 [#############...........] mikedb.sgmplus 4.72GB/8.41GB (56.2%) 2017-11-26T16:37:19.094+0100 [#############...........] mikedb.sgmplus 4.80GB/8.41GB (57.1%) 2017-11-26T16:37:22.091+0100 [#############...........] mikedb.sgmplus 4.88GB/8.41GB (58.0%) 2017-11-26T16:37:25.093+0100 [##############..........] mikedb.sgmplus 4.96GB/8.41GB (59.0%) 2017-11-26T16:37:28.094+0100 [##############..........] mikedb.sgmplus 5.04GB/8.41GB (60.0%) 2017-11-26T16:37:31.092+0100 [##############..........] mikedb.sgmplus 5.12GB/8.41GB (60.9%) 2017-11-26T16:37:34.091+0100 [##############..........] mikedb.sgmplus 5.20GB/8.41GB (61.8%) 2017-11-26T16:37:37.092+0100 [###############.........] mikedb.sgmplus 5.28GB/8.41GB (62.8%) 2017-11-26T16:37:40.094+0100 [###############.........] mikedb.sgmplus 5.36GB/8.41GB (63.7%) 2017-11-26T16:37:43.091+0100 [###############.........] mikedb.sgmplus 5.44GB/8.41GB (64.7%) 2017-11-26T16:37:46.093+0100 [###############.........] mikedb.sgmplus 5.52GB/8.41GB (65.7%) 2017-11-26T16:37:49.093+0100 [###############.........] mikedb.sgmplus 5.60GB/8.41GB (66.6%) 2017-11-26T16:37:52.091+0100 [################........] mikedb.sgmplus 5.68GB/8.41GB (67.5%) 2017-11-26T16:37:55.093+0100 [################........] mikedb.sgmplus 5.76GB/8.41GB (68.5%) 2017-11-26T16:37:58.093+0100 [################........] mikedb.sgmplus 5.84GB/8.41GB (69.4%) 2017-11-26T16:38:01.090+0100 [################........] mikedb.sgmplus 5.92GB/8.41GB (70.4%) 2017-11-26T16:38:04.095+0100 [#################.......] mikedb.sgmplus 6.00GB/8.41GB (71.3%) 2017-11-26T16:38:07.094+0100 [#################.......] mikedb.sgmplus 6.08GB/8.41GB (72.3%) 2017-11-26T16:38:10.093+0100 [#################.......] mikedb.sgmplus 6.16GB/8.41GB (73.2%) 2017-11-26T16:38:13.093+0100 [#################.......] mikedb.sgmplus 6.23GB/8.41GB (74.1%) 2017-11-26T16:38:16.093+0100 [##################......] mikedb.sgmplus 6.31GB/8.41GB (75.0%) 2017-11-26T16:38:19.091+0100 [##################......] mikedb.sgmplus 6.39GB/8.41GB (76.0%) 2017-11-26T16:38:22.091+0100 [##################......] mikedb.sgmplus 6.46GB/8.41GB (76.9%) 2017-11-26T16:38:25.094+0100 [##################......] mikedb.sgmplus 6.54GB/8.41GB (77.8%) 2017-11-26T16:38:28.091+0100 [##################......] mikedb.sgmplus 6.62GB/8.41GB (78.7%) 2017-11-26T16:38:31.093+0100 [###################.....] mikedb.sgmplus 6.70GB/8.41GB (79.6%) 2017-11-26T16:38:34.094+0100 [###################.....] mikedb.sgmplus 6.77GB/8.41GB (80.6%) 2017-11-26T16:38:37.094+0100 [###################.....] mikedb.sgmplus 6.85GB/8.41GB (81.4%) 2017-11-26T16:38:40.092+0100 [###################.....] mikedb.sgmplus 6.92GB/8.41GB (82.4%) 2017-11-26T16:38:43.092+0100 [###################.....] mikedb.sgmplus 7.00GB/8.41GB (83.2%) 2017-11-26T16:38:46.093+0100 [####################....] mikedb.sgmplus 7.08GB/8.41GB (84.2%) 2017-11-26T16:38:49.092+0100 [####################....] mikedb.sgmplus 7.15GB/8.41GB (85.1%) 2017-11-26T16:38:52.093+0100 [####################....] mikedb.sgmplus 7.23GB/8.41GB (86.0%) 2017-11-26T16:38:55.092+0100 [####################....] mikedb.sgmplus 7.31GB/8.41GB (86.9%) 2017-11-26T16:38:58.093+0100 [#####################...] mikedb.sgmplus 7.38GB/8.41GB (87.8%) 2017-11-26T16:39:01.091+0100 [#####################...] mikedb.sgmplus 7.46GB/8.41GB (88.7%) 2017-11-26T16:39:04.092+0100 [#####################...] mikedb.sgmplus 7.54GB/8.41GB (89.6%) 2017-11-26T16:39:07.093+0100 [#####################...] mikedb.sgmplus 7.61GB/8.41GB (90.5%) 2017-11-26T16:39:10.092+0100 [#####################...] mikedb.sgmplus 7.69GB/8.41GB (91.4%) 2017-11-26T16:39:13.092+0100 [######################..] mikedb.sgmplus 7.77GB/8.41GB (92.4%) 2017-11-26T16:39:16.093+0100 [######################..] mikedb.sgmplus 7.84GB/8.41GB (93.2%) 2017-11-26T16:39:19.100+0100 [######################..] mikedb.sgmplus 7.92GB/8.41GB (94.2%) 2017-11-26T16:39:22.100+0100 [######################..] mikedb.sgmplus 8.00GB/8.41GB (95.1%) 2017-11-26T16:39:25.095+0100 [#######################.] mikedb.sgmplus 8.07GB/8.41GB (96.0%) 2017-11-26T16:39:28.096+0100 [#######################.] mikedb.sgmplus 8.15GB/8.41GB (96.9%) 2017-11-26T16:39:31.100+0100 [#######################.] mikedb.sgmplus 8.22GB/8.41GB (97.8%) 2017-11-26T16:39:34.100+0100 [#######################.] mikedb.sgmplus 8.30GB/8.41GB (98.7%) 2017-11-26T16:39:37.098+0100 [#######################.] mikedb.sgmplus 8.38GB/8.41GB (99.7%) 2017-11-26T16:39:38.187+0100 [########################] mikedb.sgmplus 8.41GB/8.41GB (100.0%) 2017-11-26T16:39:38.187+0100 imported 10000000 documents Michaels-MBP:json sgm+ mdo$
Look at that, 10 million samples, most containing SGM Plus profiles, imported in 6 or so minutes on a little MacBook Pro (some basic specs below)!
Michaels-MBP:json sgm+ mdo$
Michaels-MBP:json sgm+ mdo$
Michaels-MBP:json sgm+ mdo$
Michaels-MBP:json sgm+ mdo$ /usr/sbin/system_profiler SPHardwareDataType
Hardware:
Hardware Overview:
Model Name: MacBook Pro
Model Identifier: MacBookPro14,3
Processor Name: Intel Core i7
Processor Speed: 2,9 GHz
Number of Processors: 1
Total Number of Cores: 4
L2 Cache (per Core): 256 KB
L3 Cache: 8 MB
Memory: 16 GB
Boot ROM Version: MBP143.0167.B00
SMC Version (system): 2.45f0
Serial Number (system): ***************
Hardware UUID: *********************
That is not too shabby a performance for a bit of exploratory work done without that much thought while sitting at a kitchen table on a Sunday afternoon 🙂 Admittedly it is a local database connection too, and I am just saying this is a sizable bit of single data (ca. 8.5GB) and MongoDB is not showing signs of choking at this early stage.
For performance reasons, lets slap on a few indexes. There is nothing fancy here; to efficiently retrieve a known Sample ID, an index on Sample ID is necessary, to efficiently retrieve details for a specific Extraction plate and well location, a composite index is necessary there too. The following few lines show typical index creation.
db.sgmplus.ensureIndex({"sampleID":1}, { background: true } )
db.sgmplus.ensureIndex({plateNumberEXT:1} )
db.sgmplus.ensureIndex({plateNumberQUANT:1} )
db.sgmplus.ensureIndex({plateNumberPCR:1} )
db.sgmplus.ensureIndex({plateNumberCE:1} )
db.sgmplus.ensureIndex({wellLocationEXT:1} )
db.sgmplus.ensureIndex({wellLocationQUANT:1} )
db.sgmplus.ensureIndex({plateNumberPCR:1} )
db.sgmplus.ensureIndex({plateNumberCE:1} )
db.sgmplus.ensureIndex({plateNumberEXT:1, wellLocationEXT:1} )
db.sgmplus.ensureIndex({plateNumberQUANT:1, wellLocationQUANT:1} )
db.sgmplus.ensureIndex({plateNumberPCR:1, plateNumberPCR:1} )
db.sgmplus.ensureIndex({plateNumberCE:1, plateNumberCE:1} )
db.sgmplus.ensureIndex({"profile.locus":1, "profile.alleles":1})
db.sgmplus.ensureIndex({plateNumberCE:1, "profile.locus":1, "profile.alleles":1})
db.sgmplus.ensureIndex({plateNumberCE:1, wellLocationCE:1, "profile.locus":1, "profile.alleles":1})
Am I flippantly dismissing indexing by, specifically, not providing too much detail. Yes, and for a deliberate reason, so keep reading. However, with the indexes above, each of the database queries below is satisfied with (sub-)second performance, with the exception of query 06 where the query response time was around 6 seconds..
Now, if you have made it thus far, the point of the preamble above is to paint the picture, create some not unreasonable test data, and now build and execute some database queries required in a typical DNA forensics service provider. Of course the whole point is to establish whether MongoDB is up to the job of being rolled as the technology of choice in a SGMPlus (or other multiplex) DNA database.
Some typical queries might be:
Database Query 01: How many samples details are present in the database
> > db.sgmplus.count(); 10000000 > >
Database Query 02: Of all the sample (details) that are present in the database, how many have SGMPlus DNA profiles associated with them.
The three ways that this could be achieved are:
- an aggregation pipeline
- a single purpose aggregation operation, eg. count, sum, ….
- map-reduce
I demonstrate application of the former two options below. Map-reduce usage is demonstrated vide infra.
>
>
>
> db.sgmplus.find({profile : {$exists : true}}).count()
7696545
>
>
>
> db.sgmplus.aggregate(
... [
... { $match : { profile : {$exists : true} } },
... { $group : { _id: null, total : { $sum : 1 } } },
... { $project : { _id : 0 } }
... ]
... ).pretty()
{ "total" : 7696545 }
>
>
>
>
Database Query 03: Retrieve all the laboratory workflow data available for sampleID 2, excluding the DNA profile, and Quantitation plate ID and well location
>
>
>
>
>
> db.sgmplus.find({sampleID:2}, {_id:0, plateNumberQUANT:0, wellLocationQUANT:0, profile:0}).pretty();
{
"sampleID" : 2,
"plateNumberEXT" : "E00032461",
"wellLocationEXT" : "D06",
"plateNumberPCR" : "P00021200",
"wellLocationPCR" : "D02",
"plateNumberCE" : "C00017800",
"wellLocationCE" : "E12",
"caseID" : 171162
}
>
>
>
Database Query 04: Retrieve just the extraction plate number and well location for the sample found on PCR plate number P00021200 well location D02
>
>
>
> db.sgmplus.find( { plateNumberPCR : "P00021200", wellLocationPCR : "D02" }, { plateNumberEXT : 1, wellLocationEXT : 1, _id : 0} );
{ "plateNumberEXT" : "E00032461", "wellLocationEXT" : "D06" }
>
>
>
Database Query 05: Aneuploidy (aneuploidy is not a service of interest in forensic DNA typing, but for the purposes of MongoDB evaluation, and also querying the database for specific loci and allele inclusion/exclusion criteria, it is a good test case. It also piques my interest, so why not confirm whether MongoDB is up to the job here too).
05a: Of all the DNA profiles present, how many XXX women (so excluding XXF) are present
>
>
>
> db.sgmplus.find( {
... profile : {
... $elemMatch : { locus : "Amel", alleles : { $size : 3, $nin : ["F", "Y"] } }
... }
... }
... ).count()
23356
>
>
>
05b: Of all the DNA profiles present, retrieve an SGMPlus profile of a sample that exhibits Edward (trisomy 18) syndrome
>
>
>
>
> db.sgmplus.findOne( {
... profile : {
... $elemMatch : { locus : "D18", alleles : { $size : 3 } }
... }
... }
... );
{
"_id" : ObjectId("59fc51d21f11734851d20b24"),
"sampleID" : 764,
"plateNumberEXT" : "E00032469",
"wellLocationEXT" : "C12",
"plateNumberQUANT" : "Q00005507",
"wellLocationQUANT" : "G04",
"plateNumberPCR" : "P00021201",
"wellLocationPCR" : "U04",
"plateNumberCE" : "C00017801",
"wellLocationCE" : "R15",
"profile" : [
{
"locus" : "D21",
"alleles" : [
"28.1"
]
},
{
"locus" : "D18",
"alleles" : [
"10",
"11",
"21.2"
]
},
{
"locus" : "TH01",
"alleles" : [
"8.1",
"6.3"
]
},
{
"locus" : "D16",
"alleles" : [
"7",
"11.2"
]
},
{
"locus" : "D19",
"alleles" : [
"F",
"14"
]
},
{
"locus" : "vWA",
"alleles" : [
"20",
"24"
]
},
{
"locus" : "D2",
"alleles" : [
"26",
"16.2"
]
},
{
"locus" : "FGA",
"alleles" : [
"48",
"25.2"
]
},
{
"locus" : "Amel",
"alleles" : [
"F",
"F"
]
},
{
"locus" : "D8",
"alleles" : [
"9.2",
"16.2"
]
},
{
"locus" : "D3",
"alleles" : [
"18.2",
"12.2"
]
}
],
"caseID" : 883614
}
>
>
>
>
>
>
>
05c: Of all the DNA profiles present, retrieve an SGMPlus profile of an XXX women that have an explicit allelic designation of 15 and 16 for TH01 (diploid)
>
>
>
>
> db.sgmplus.findOne( {
... profile : {
... $all : [
... { "$elemMatch" : { locus : "Amel", alleles : { $size : 3, $nin : ["F", "Y"] } } },
... { "$elemMatch" : { locus : "TH01", alleles : { $size : 2, $all : ["4", "5.1"] } } }
... ]
... }
... }
... );
{
"_id" : ObjectId("59fc51d31f11734851d2984a"),
"sampleID" : 36862,
"plateNumberEXT" : "E00032845",
"wellLocationEXT" : "A03",
"plateNumberQUANT" : "Q00005865",
"wellLocationQUANT" : "B01",
"plateNumberPCR" : "P00021282",
"wellLocationPCR" : "F03",
"plateNumberCE" : "C00017873",
"wellLocationCE" : "V04",
"profile" : [
{
"locus" : "D2",
"alleles" : [
"25.2"
]
},
{
"locus" : "D19",
"alleles" : [
"F",
"15"
]
},
{
"locus" : "D8",
"alleles" : [
"11.2",
"F"
]
},
{
"locus" : "D18",
"alleles" : [
"10",
"23"
]
},
{
"locus" : "D21",
"alleles" : [
"27",
"32.2"
]
},
{
"locus" : "FGA",
"alleles" : [
"32",
"26.2"
]
},
{
"locus" : "D3",
"alleles" : [
"9",
"10"
]
},
{
"locus" : "TH01",
"alleles" : [
"5.1",
"4"
]
},
{
"locus" : "vWA",
"alleles" : [
"10",
"18.2"
]
},
{
"locus" : "Amel",
"alleles" : [
"X",
"X",
"X"
]
},
{
"locus" : "D16",
"alleles" : [
"6.2",
"14.2",
"14",
"5"
]
}
],
"caseID" : 714981
}
>
>
>
>
>
05d: How many examples of pentasomy exist in the database (note may be more than one example in an SGMPlus profile per sample). As above, note this is randomly generated data. This type of chromosomal abnormality is exceptionally rare.
>
>
> db.sgmplus.find( { "profile.alleles" : { $size : 5 } } ).count();
681
>
>
05e: How many discrete samples in the SGMPlus test database exhibit pentasomy
>
>
>
> db.sgmplus.distinct("sampleID", { "profile.alleles" : { $size : 5 } }).length;
679
>
>
Database Query 06: Retrieve the SGMPlus profile on CE/Gel number C00017873 well V03 and display the allelic values sorted numerically lowest to highest, with Fail (F) last and for designation with amelogen X precedes Y. Separate the alleles by commas, surround by (), prefix with the loci, and separate each loci/allele list by a semicolon (making the DNA profile human-readable, but also easily digestible by C$WILDNA1, an Oracle Data Cartridge that enhances Oracle so that it may efficiently index DNA profiles on-a-par with Oracle native data types).
An example profile is shown below:
D8 (16.2,20); D21 (31,36.2); FGA (28.2,51.2); D18 (25,F); D3 (14,18.2); TH01 (9.1); D2 (21.2,27.2); Amel (X,Y,Y); D16 (4,9.2); vWA (15.2,20.2); D19 (10.2,15.2);
An SGMPlus profile rendered in this C$WILDNA1 and human-digestible format is shown below.
>
>
>
>
>
>
>
>
> db.sgmplus.mapReduce(
... function(){
... var csv = "";
... for (i = 0; i < this.profile.length; i++)
... {
... var locus = this.profile[i]["locus"]; //TH01, D21, Amel etc.
... var alleles = this.profile[i]["alleles"];
...
... var sortedAlleles = alleles.sort(
... function(a, b)
... {
... var aNaN = isNaN(a);
... var bNaN = isNaN(b);
...
... if(aNaN && bNaN)
... { // X before Y, and F last regardless
... if (a == 'F')
... return true;
... if (b == 'F')
... return false;
... return a > b;
... }
...
... if (!aNaN && !bNaN)
... {
... return Number(a) > Number(b);
... }
...
... //one is alphabetic, one numeric, and allelic numbers come before the "F" (Fail)
... return aNaN;
... }
... );
...
... csv = csv + locus + " (" + sortedAlleles + "); ";
... }
...
... emit({_id : this.sampleID}, { "sortedFormattedSGMplusProfile" : csv});
... },
... function() {},
... {
... query : { plateNumberCE : "C00017873", wellLocationCE : "V06" },
... out: { inline : 1 }
... }
... ).find();
[
{
"_id" : {
"_id" : 36864
},
"value" : {
"sortedFormattedSGMplusProfile" : "D8 (16.2,20); D21 (31,36.2); FGA (28.2,51.2); D18 (25,F); D3 (14,18.2); TH01 (9.1); D2 (21.2,27.2); Amel (X,Y,Y); D16 (4,9.2); vWA (15.2,20.2); D19 (10.2,15.2); "
}
}
]
>
>
>
This requirement initially appeared to be trivial, however when you are still honing your MongoDB skills (as I am), things quickly ballooned into the more complex query above. Regardless of the apparent complex formatting requirement, that can also easily be done in middleware C# code, or another programming language, the requirement to render DNA profiles in a manner to something that resembles CSV is a common and very valid requirement in DNA typing laboratories.
Database Query 07: Laboratory QA, QC, Accreditation, and BI purposes
7a. A rape victims DNA profile was determined from a sample located on CE/Gel plate number C00017908 well location E12. The rapist/suspects DNA sampleID is 53837. At any point in any part of the laboratory workflow (excluding Quantitation) did a plate ever contain both sample extracts from both the rapist/suspect and the rape victim.
>
>
>
>
>
> //
> // The query below will return 1 if extracts from the victim and the suspect/rapist were ever on the same
> // laboratory plate, or 0 otherwise
> //
> db.sgmplus.aggregate(
... [
... { $match : { plateNumberCE : "C00017908" } },
... { $lookup:
... {
... from : "sgmplus",
... localField: "plateNumberEXT",
... foreignField: "plateNumberEXT",
... as: "priorEXT"
... }
... },
... { $lookup:
... {
... from : "sgmplus",
... localField: "plateNumberPCR",
... foreignField: "plateNumberPCR",
... as: "priorPCR"
... }
... },
... { $lookup:
... {
... from : "sgmplus",
... localField: "plateNumberCE",
... foreignField: "plateNumberCE",
... as: "priorCE"
... }
... }
... , { $project : { sampleIDs : ["$priorEXT.sampleID", "$priorPCR.sampleID", "$priorCE.sampleID" ] } }
... , { $unwind : '$sampleIDs' } //unwind twice, nested array
... , { $unwind : '$sampleIDs' }
... , { $group : { _id : 0,
... allSampleIDs : { $addToSet : "$sampleIDs"}
... }
... }
... , { $match : { allSampleIDs : 53837 } }
... ]
... ).itcount()
1
>
>
>
>
7b. Determine the unique set of all prior plates that ultimately had samples that ended up in a well on CE/Gel plate C00017908 used to derive the rape victims DNA profile (question 7a above).
>
>
>
>
>
>
> db.sgmplus.aggregate(
... [
... { $match : { plateNumberCE : "C00017908" } },
... { $project : { allPlates : [ "$plateNumberPCR", "$plateNumberCE", "$plateNumberQUANT", "$plateNumberEXT" ] } },
... { $unwind : '$allPlates' },
... { $group : { _id : 0,
... priorLaboratoryWorkflowPlateNumbers : { $addToSet : "$allPlates"}
... }
... }
... ]
... ).pretty()
{
"_id" : 0,
"priorLaboratoryWorkflowPlateNumbers" : [
"P00021321",
"E00033026",
"E00033025",
"Q00006036",
"P00021319",
"Q00006035",
"P00021320",
"E00033023",
"Q00006032",
"E00033022",
"C00017908",
"Q00006033",
"Q00006037",
"E00033021",
"E00033024",
"Q00006034"
]
}
>
>
>
>
Database Query 08: DNA Profile Hit, Near and Partial Matches
- Does an exact match exist in the database for a SGMPlus DNA profile, ie. a hit: we have the DNA of the rapist/suspect and does it match that found on/in the victim
- Does a near or partial match exist in the database for a SGMPlus DNA profile, ie. a likely hit, a familial match – the DNA isn’t that of the rapist, but perhaps a (half) brother, uncle, or other relative (or even contamination – a staff member in the laboratory has contaminated the sample or plates, a police officer has deposited their database at the crime scene and it has been submitted for analysis, or even intra-sample contamination within the laboratory workflow).
Examples of these queries are not shown herein, and the source code for the database queries is also a little more complex. Furthermore for performance reasons, judicious use of database indexes is critical (note throw-away comments and specific lack of detail on indexes above). A second blog post dedicated to these two specific queries will soon be written, they are worthy of their own blog article and this blog post is already too long …….. stay tuned.
Summary
MongoDB doesn’t even break into a sweat when 10,000,000 bits of sample tracking information are added to a single collection object and queried. In the PoC, queries typical of a forensic laboratory (and then some) were executed solely for SGMPlus, but could easily be applied to other multiplex kits. Database performance is good (certainly comparable or better than the COTS informatics LIMS implementations I have seen in a number of forensic service providers) and the query language rich and where necessary, a splash of Javascript yields the required results (eg. database query 06 above). The performance findings herein qualitative, despite my best intentions to include quantitative information when I started writing this article. A second blog article will follow with quantitative performance/timing metrics and MongoDB examples of exact, near, and partial SGMPlus DNA profile matching. For informatics practitioners and forensic scientists reading this blog, note I am fully aware of the implications of non-concordance across profiling kits, and “versions” of the same kit. This subject matter will be specifically addressed in another blog post. Put another way, two DNA profiles that exactly match are not necessarily a genetic match/database hit, despite what some people in IT that write this type of DNA profile matching software may think!
The Perl code for SGMPlus example data generation can be downloaded from GitHub.
— Published by Mike, 12:45:41 30 November 2017 (CET)
Archive
By Month: November 2022, October 2022, August 2022, February 2021, January 2021, December 2020, November 2020, March 2019, September 2018, June 2018, May 2018, April 2018
Categories
Apple, C#, Databases, Faircom, General IT Rant, German, Informatics, LINQ, MongoDB, Oracle, Perl, PostgreSQL, SQL, SQL Server, Unit Testing, XML/XSLT



Leave a Reply